AI intraoperative classification of central nervous system tumors based on simulated nanopore sequencing
J. de Ridder and colleagues conducted a study on the development and use of a neural network classifier called Sturgeon for classifying central nervous system (CNS) tumors during surgery. They have recently published their work in Nature (DOI:10.1038/s41586-023-06615-2). The team addressed the limitations of current methods for determining precise tumor types prior to surgery and explored the potential benefits of using rapid nanopore sequencing to obtain a methylation profile during surgery. The Sturgeon classifier, optimized to handle sparse data, was trained on simulated nanopore sequencing runs. The study demonstrated that Sturgeon could accurately classify CNS tumor samples based on as little as 20-40 minutes of sequencing time. In a realistic intraoperative setting, Sturgeon showcased its ability to influence surgical decision-making by providing a diagnostic turnaround time of less than 90 minutes. The importance of a precise and reliable diagnosis for guiding neurosurgical strategies was emphasized. The use of nanopore sequencing offered advantages such as low setup cost, small form factor, and instant data availability. Although the Sturgeon approach required a certain amount of tissue, successful tests with smaller samples suggested it could potentially assist in preventing neurological comorbidity and the need for additional surgeries.
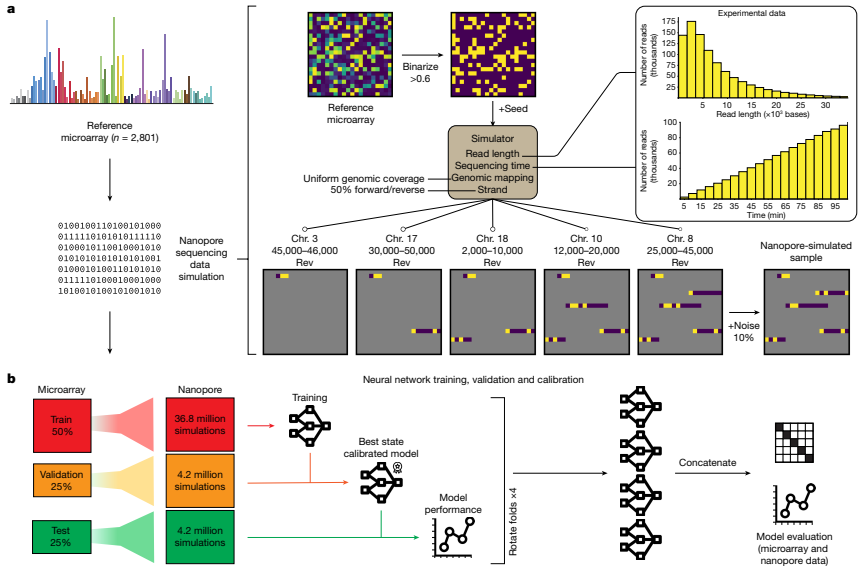
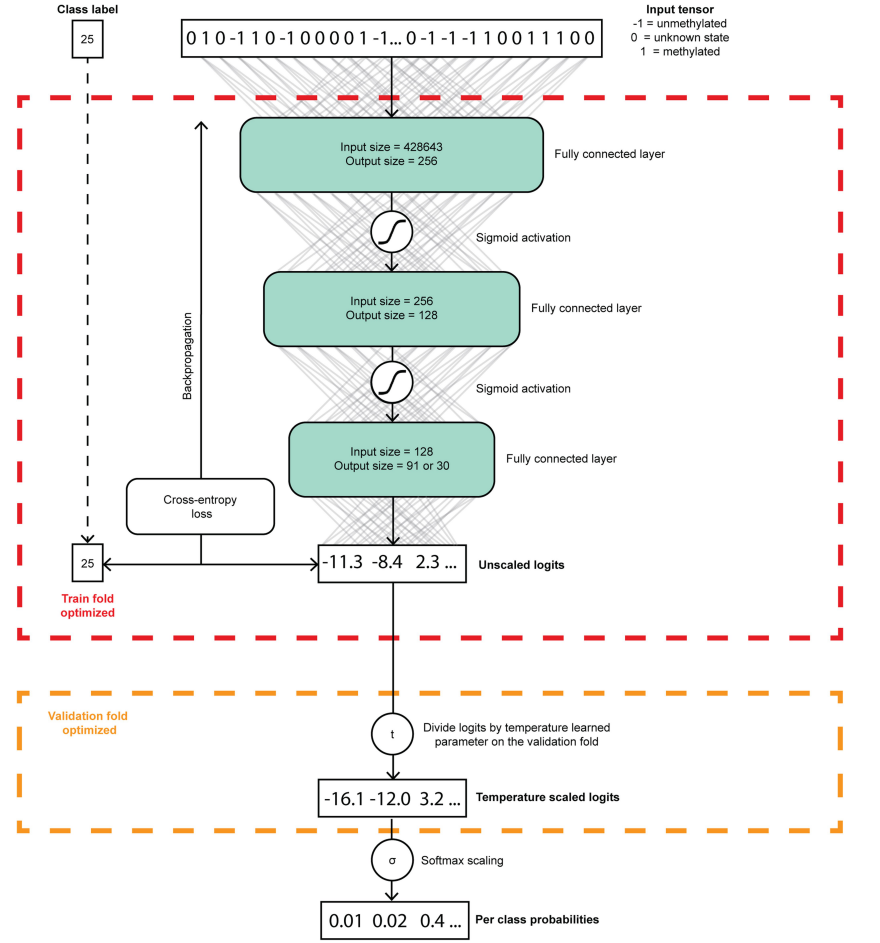
The researchers used simulated nanopore sequencing data to train and verify their Sturgeon model for CNS tumor classification. Specifically, they used four neural networks that were each trained, validated, and calibrated independently. The training data consisted of simulations generated from the Capper et al. reference dataset, which was split into four folds while maintaining the original class distributions. The researchers used two folds to train the submodel, one fold to determine the best-performing state of the submodel and perform score calibration, and the final fold to evaluate the submodel’s performances. The neural networks were pre-trained using simulations that ranged from 0.6% to 14% sparsity, and then fine-tuned for the final classifier using simulations that ranged from 0.6% to 6.3% sparsity. To verify the model’s performance, the researchers assessed its final performance on a left-out test fold, with each sample contributing 6,000 simulated samples to the test set. They evaluated the classification accuracy and other metrics across all sparsity levels to evaluate the model’s performance. The researchers also validated their model on pediatric methylation profiles obtained from routine diagnostic processes. Additionally, they performed classification of publicly available nanopore sequencing data obtained from GSE209865. In short, the Sturgeon model was trained and verified using simulated nanopore sequencing data, including simulations from the Capper et al. reference dataset and publicly available nanopore sequencing data. The model’s performance was evaluated using various metrics and validated on real pediatric methylation profiles.
The training dataset for Sturgeon consists of a varied population of patients of different ages (mean age of 29 with 36% less than 13 years of age). We first aimed to further validate the performance of Sturgeon in a paediatric setting. For this purpose, we obtained 94 genome-wide methylation profiles generated using the Illumina Infinium Methyla-tionEPIC v1.0 (hereafter referred to as EPIC) arrays from patients that underwent a CNS tumour resection surgery in the Princess Máxima Center (PMC) for paediatric oncology. For each of these samples, the publicly available Heidelberg classifier (v11b4) was applied as part of the routine clinical care. This classifier can be considered an updated version of the Capper et al. classifier. The recommended cut-off for a clinical diagnosis is 0.84, which the classifier reached for the majority (n = 68) of samples. Those classified below the 0.84 cut-off are considered difficult to diagnose based on their methylation pro-file, which is likely to occur for uncommon tumour types that do not correspond to any of the previously annotated classes, tumours that occur in the context of a genetic tumour predisposition syndrome, heterogeneous samples or samples with a low tumour purity.For each methylation profile we simulated 500 nanopore sequenc-ing experiments at 7 sequencing depths for a total of 332,500 simulated nanopore sequencing experiments, after which we applied the Sturgeon classifier
Neurosurgeons can utilize the Sturgeon neural network classifier during surgery to predict tumor classification. The Sturgeon classifier is based on rapid nanopore sequencing, which allows for the acquisition of a methylation profile of the tumor during the surgical procedure. This method overcomes the limitations of current preoperative imaging and intraoperative histological analysis, which may not always provide a conclusive or accurate tumor type diagnosis.
The Sturgeon classifier is optimized to handle sparse data and has been trained on simulated nanopore sequencing runs . It can accurately classify central nervous system (CNS) tumor samples based on as little as 20-40 minutes of sequencing. In a realistic intraoperative setting, Sturgeon has demonstrated the ability to provide a diagnostic turnaround time of less than 90 minutes, influencing surgical decision-making. By rapidly obtaining a molecular subtype diagnosis, neurosurgeons can make more informed decisions regarding the extent of tumor resection, minimizing the risk of neurological damage and comorbidity .
The use of nanopore sequencing in the Sturgeon approach offers advantages such as low setup cost, a small form factor, and instant data availability. Although a certain amount of tissue is currently required for the Sturgeon approach, it has been successfully tested with smaller samples. Overall, utilizing the Sturgeon classifier during surgery can assist neurosurgeons in achieving a precise and reliable tumor classification, guiding their surgical strategies and potentially preventing neurological comorbidity and the need for additional surgeries.